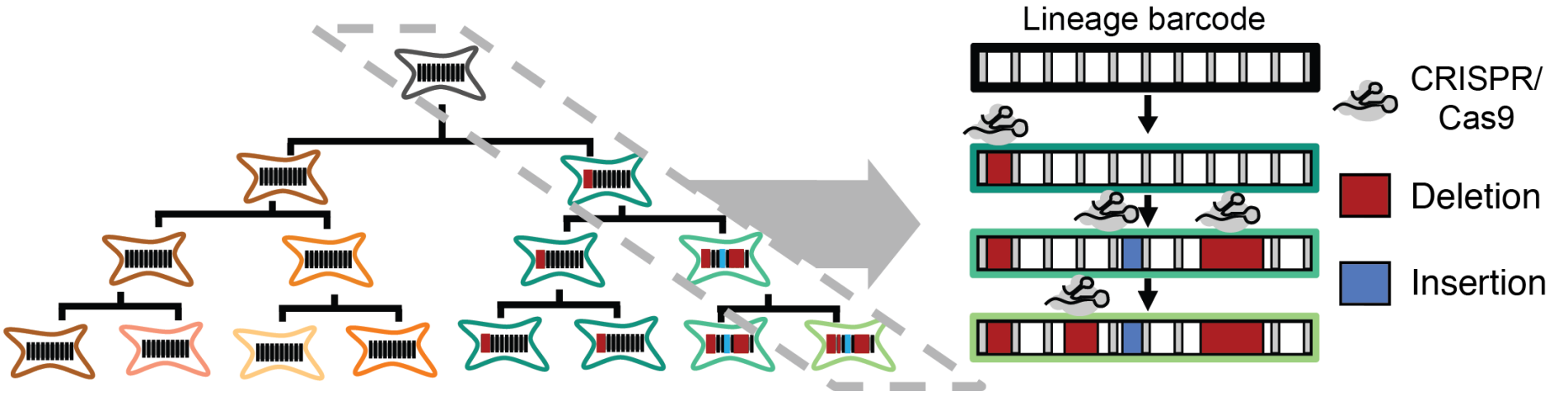
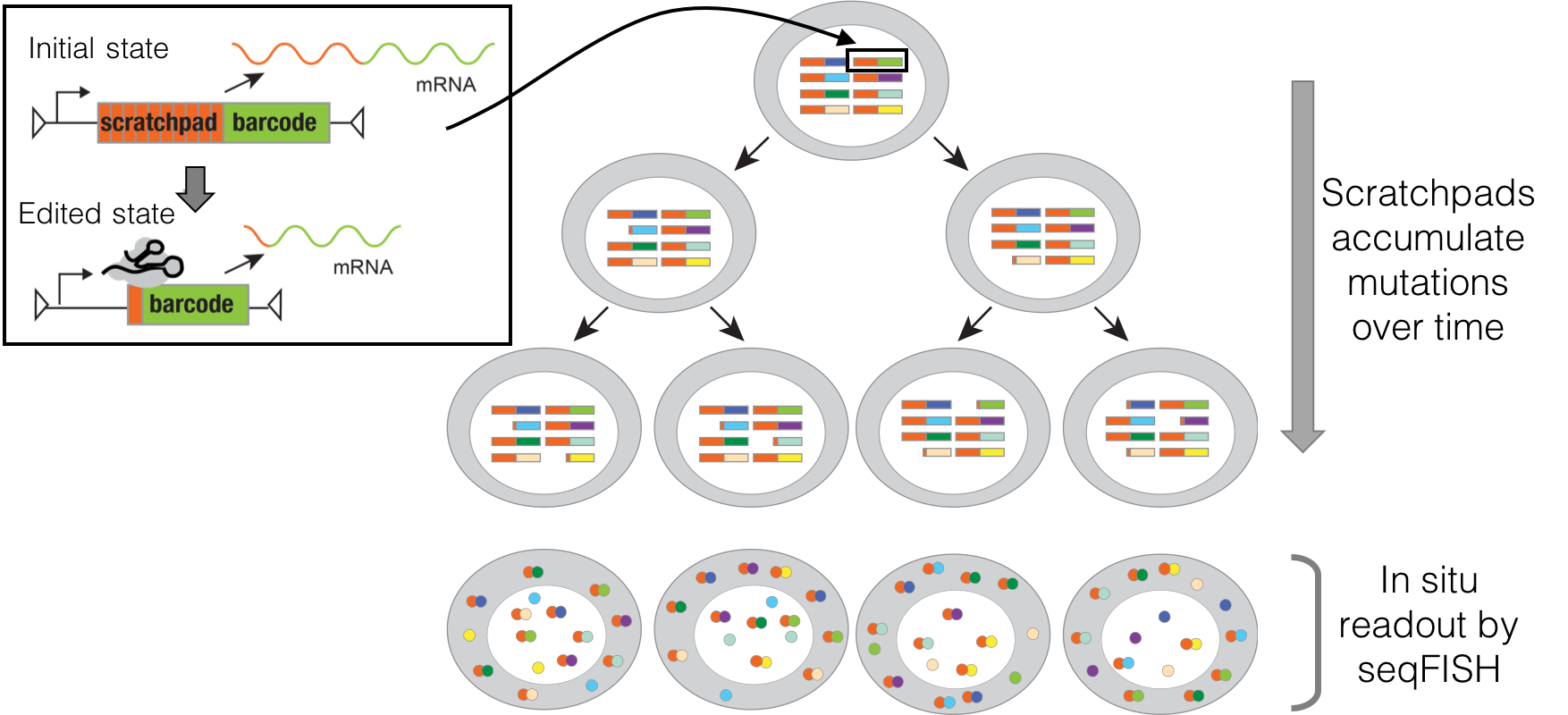
We each began as one cell, which divided into two, and then into four, eight, sixteen and so on, ultimately giving rise to the highly organized mass of forty trillion cells that constitutes an adult human. A fundamental scientific question is to understand the process by which this family tree or “lineage” of cells unfolds in space and time. How is each cell related to each other cell? How closely are different kinds of cells related? Who and where were their common ancestors? What are the signals that drove their differentiation into different types? When and from whom were these signals received? For over a century, biologists have been asking these questions, but our understanding of how humans and other vertebrates develop remains highly incomplete. Over the past few years, our team pioneered a novel paradigm for globally recording the histories of cells. In brief, we use a powerful new genome editing tool, CRISPR, to irreversibly record information by mutating targeted regions of the genome during an organism’s development. Because the genome is inherited by each daughter of a dividing cell, the accumulated mutations can be subsequently read out and used to computationally reconstruct the lineage relationships between cells of different types and locations, as well as to recover additional aspects of their molecular histories. The Allen Discovery Center for Cell Lineage Tracing is developing and applying this paradigm in two key models for human biology: the zebrafish and mouse. We anticipate that the resulting global maps of development will be broadly transformative, with major impacts across developmental biology, neuroscience, cancer biology, regenerative medicine, and other fields, opening up new areas of inquiry, settling long-standing questions, and serving as a fundamental resource for our collective efforts to understand the remarkable complexity and robustness of development in humans and other vertebrates.