DREAM Challenge
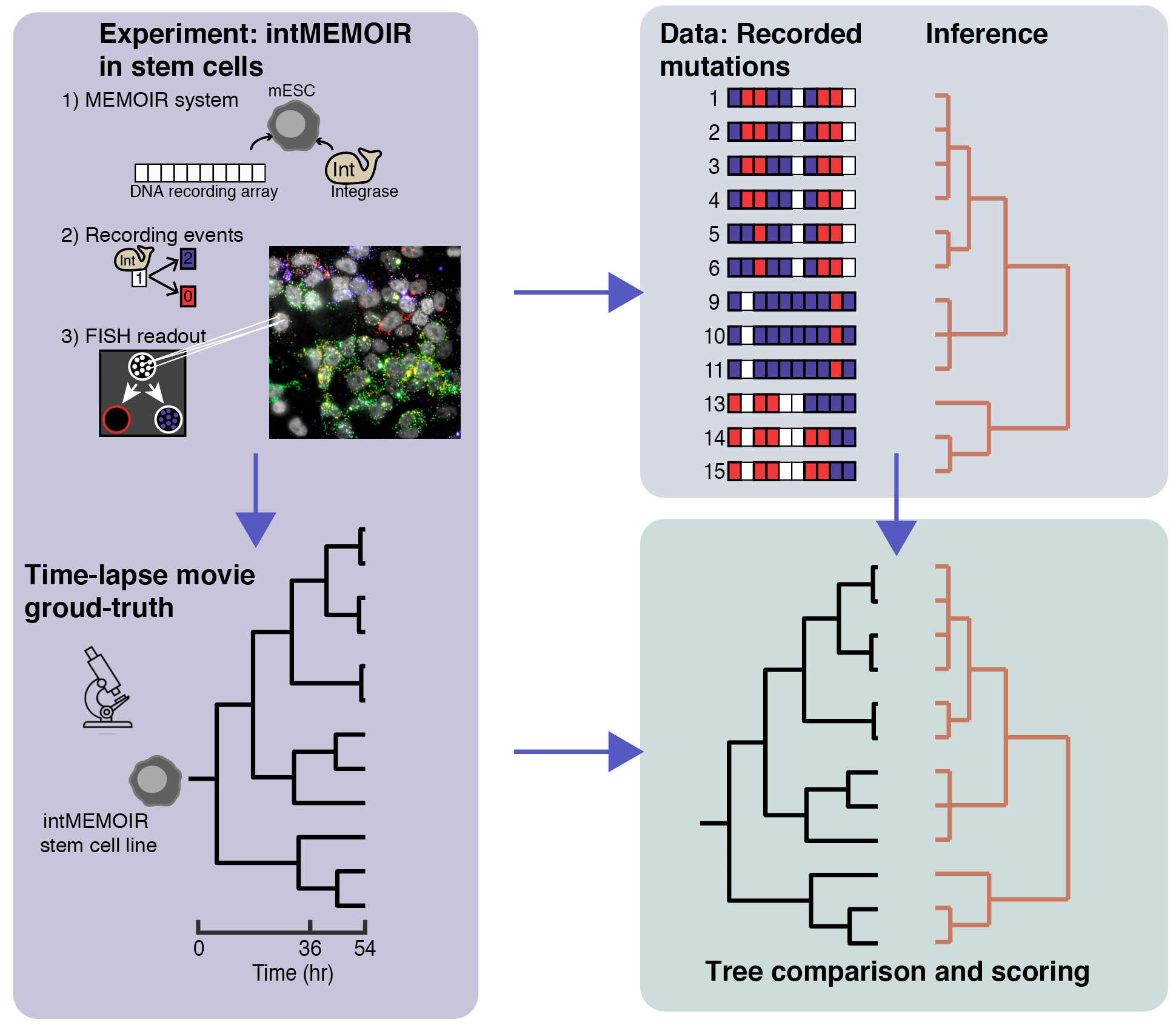
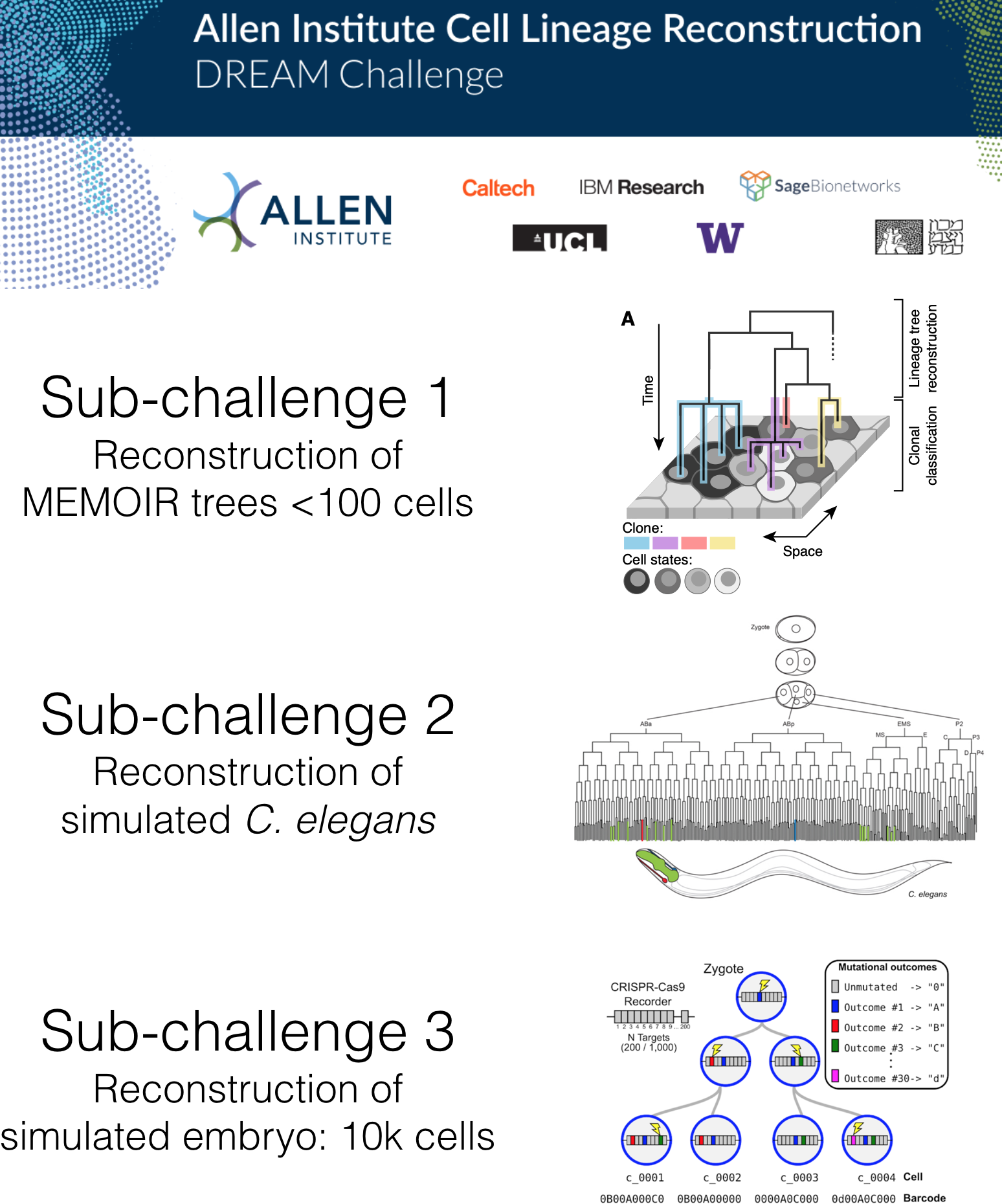
To catalyze the development of new methods to perform lineage reconstruction, we organized the Allen Institute Lineage Reconstruction DREAM Challenge, which ran from October 2019 through February 2020. DREAM challenges are a platform for crowdsourcing collaborative competitions where a rigorous evaluation of each submitted solution allows for objective comparison and assessment of their performance. The value of DREAM resides not only in the acceleration of research through the participation of many teams in solving a problem, but just as importantly, in the diversification of approaches used in emerging areas of biology and in the quality and reproducibility of each provided solution.
Github repositories for examples of the best performing challenge methods are linked below.
Sub-challenge 1 winners: Cassiopeia (Yosef Lab)
Cassiopeia is a software suite for processing data from single cell lineage tracing experiments. This suite comes equipped with three main modules: Target Site Sequencing Pipeline, Phylogeny Reconstruction, and Benchmarking.
https://github.com/YosefLab/Cassiopeia
Sub-challenges 2 and 3 winners: Distance based Cell LinEAge Reconstruction (DCLEAR) (Il-Youp Kwak and Wuming Gong)
R/DCLEAR is an R package for Distance based Cell LinEAge Reconstruction(DCLEAR).
https://github.com/ikwak2/DCLEAR
Additional information about the Allen Discovery Center for Lineage Tracing DREAM Challenges is available here:
http://dreamchallenges.org/project/allen-institute-cell-lineage-reconstruction-dream-challenge/